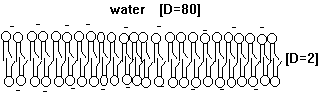|
The Development of a Scientific Fact
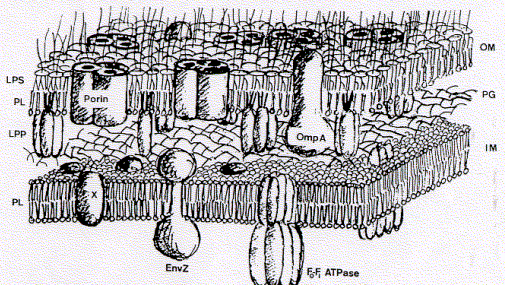
What the model says The fluid mosaic model of a biological membrane defines a phospholipid bilayer mixed with (integral) membrane proteins. The lipid bilayer, therefore, is not a continuous layer, but contains non-lipid molecules, in particular proteins.
The significance of the model The fluid mosaic model is the only accepted proper description of a biological membrane. Its publication in 1972 by Singer and Nicolson (Science 1972 Feb. 18 175:23 720-31) put an end to other existing and competing models of biological membranes. These competing models assumed triple layers, with a lipid core layer and two protein layers, one on each side, but the proteins would not penetrate the lipid core, i.e., did not span the membrane. The fluid mosaic model, based on thermodynamic and functional considerations, postulated instead that proteins as the active components of signal transduction and transport, must and can span the entire thickness of a lipid bilayer. Protein-lipid interaction was thought to be stabilized through hydrophobic (fat soluble) contact points on the surface of the proteins. Today, the fluid-mosaic model is the only accepted model of the structure of biological membranes, including those of animals, plants, and microorganisms (see figure above for membranes of the cell wall of the bacteria Escherichia coli). the use of the same structure among all living organisms is consistent with the idea of a common ancestor organism (or organisms) from which all modern life forms have evolved. Since 1972 no modifications of the model have been necessary, also individual membranes differ in molecular composition, lipid types, or lipid to protein ratios. To understand the stability and form of membranes one needs to understand that they are solely determined by the aggregation behavior of phospholipids. These lipid types in effect form extensive, planar bimolecular films separating two aqueous compartments.
The hydrophobic core of such a thin film functions as an electrical insulator (D=2; D stands for dielectric constant; a value of 2 represents hydrophobic environment, a value of 80 water) allowing diffusion of small hydrophobic solutes across membranes, but inhibits the diffusion of charged and hydrophilic, i.e., water soluble molecules. The incorporation of protein into these bimolecular films provides the necessary pathways for all charged and hydrophilic molecules. Cells have thus complete control over their membrane permeability because they can control protein function, but not the diffusion of small hydrophobic molecules across membranes. Cell membranes are thus clearly specific and selective. To understand the current model of biological membranes, one has
to know the history of the study of cell membranes, both from a
structural and functional point of view. Early developments The first experimental indications for a bilayer structure have been obtained by measuring the size of a water surface that can be covered by phospholipids extracted from red blood cells. This area was about twice as large as the total surface area of red blood cells used to extract lipids. It turned out this first calculation was a lucky shot and with more accurate measurement, the two surface ratios would not have been 2:1 due to the presence of membrane proteins. This original finding, although not reproducible, stimulated membrane research and guided it into the right direction. Neurobiologist started to characterize the function of neuronal cells by measuring action potentials which are the result of electrical currents flowing across cell membranes (called excitable membranes of nerve and muscle cells). Macroscopic current recordings of isolated nerve cells in the 1940s allowed the prediction of underlying molecular units called ion channels (the electric charges are carried by ions, charged atoms and molecules). Although the sensitivity of electrical recordings was not good enough at this time to measure individual ion channels, the noise behavior of the macroscopic currents were statistically analyzed to predict the existence of unit channels. The macroscopic membrane currents were simply the results of the simultaneous activity of thousands of those unit channels. These ion channels have later been shown to be made of proteins. It was one of the great achievements for biophysicists in the 1960s to demonstrate conclusively that these current recordings can be interpreted as the opening and closing events of a single ion channel structure at a time. By the time I got to witness this experiment, more than 20 years had passed since the first membrane had ever been formed in vitro (black lipid membranes; 1962 by Mueller and Rudin), and about 12 years earlier black lipid membranes were used to study gramicidin channel activity (D.W. Urry, 1971; M.C.Goodall, 1971). The Montal & Mueller bilayer provided the only means to assemble phospholipid bilayers that emulate the asymmetric distribution of lipids found in biological membranes. These two scientists contributed an experimental system that demonstrated for the first time that purified membrane proteins could be reconstituted to full functionality and recapitulate their biological function. Their contribution was first for the nicotinic acetylcholine receptor, the voltage-gated sodium channel, the photosynthetic reaction centers, cytochrome oxidase, and later on the bacterial toxin channels, Bcl-2 family of proteins, and the HIV Vpu proteins, to name a few. The pore forming activity of gramicidin peptides had been shown
before using diverse biological membranes, such us mitochondria
and red blood cells, both depending on functional ion gradients
which are destroyed by the presence of gramicidin. Hladky and Haydon
demonstrated the unit conductance of gramicidin in 1972 which is
the functional correlate of a peptide dimer spanning a biological
(here artificial) membrane.
See table of key historic dates
for a more detailed outline of the development of the biological
membrane as a scientific fact. It is no longer up to the individual
experimentalist to speculate about the general structure of biological
membranes. For further reading on the general topic of scientific
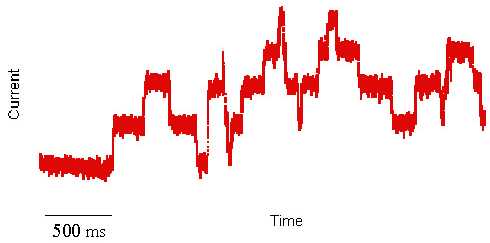
facts see Ludwik Fleck. I first heard about artificial cell membranes in the early 1980s. I was a biochemistry student at that time and considered myself a biologist. The very idea of using artificial systems to study biological processes hurt my sensibility as a young student. I did not know then that a few month later I would witness an experiment that forever changed my mind, jump-starting a scientific career devoted to membrane protein structure and function. The pivotal experiment was a two-day demonstration by a graduate student showing the mechanism of how an antibiotic compound kills bacteria by punching holes into their cell membranes. This antibiotic, called Gramicidin, is a peptide (small protein) able to kill bacteria by spontaneously inserting itself into their membranes and destroying their most important energy storing device, a proton concentration gradient. In plain English, Gramicidin kills bacteria by short circuiting their battery. What was most intriguing about this experiment I witnessed was the electrical activity that this peptide induced when inserted into an artificial membrane. By literally creating small ion (current) conducting channels across the otherwise electrically insulating cell membrane, carefully placed electrodes pick up rectangular jumps in currents exhibiting a reproducible unit conductance. Because cell membranes are extremely thin and the channel volumes very small, the electrical current has to be amplified about a billion fold. This amplification demonstrated the activity of a single molecule activity. This is not your conventional biochemical reaction, where billions of proteins are measured at once and the kinetic analysis of the chemical product formation really represent the average of every single enzyme reaction. It is impossible from such macroscopic experiments to exactly tell what's happening at a single active site. In the current recording shown above (red trace), every single rectangular transition up or down, however, does represent the activity of a single active site, i.e., the opening or closing of a transmembrane ion channel. This single protein activity can be observed only, if millions of ions flow through the channel every second. The ultimate goal to study the trajectory of a single ion through a single channel is still beyond experimental means, but can be simulated with the help of computers.
The amount of data accumulated over time by many laboratories has
set forth a body of evidence that allowed to call an experiment
like the one described here for Gramicidin D a single channel
recording beyond reasonable doubt. The functional characterization
of ion channels in biological membranes has proven the fluid mosaic
model correct. The model has become a scientific fact. It has become
a fact, even though nobody has ever observed the activity of Gramicidin
at this single molecule level in a real cell, a bacterial membrane.
It has become a fact even though nobody has ever seen a Gramicidin
peptide. Yet its molecular structure is known in great detail using
nuclear magnetic resonance, X-ray crystallography, or many other
spectroscopic techniques. It has become a fact because it was possible
to meticulously put together the pieces of a molecular puzzle into
a coherent system of observations. The structural and functional
characterization of biological membranes and its components is a
marvel of reductionist experimentation, of logic and deduction.
It is also the consequence of an astonishing property of biological
molecules -- to self assemble in aqueous solution. Copyright © 2000-2010 Lukas K. Buehler |
||||||